2.9 Regression Diagnostics
In this section we will be working with the additive analysis of covariance model of the previous section. As usual, we start by reading the data and recreating the variables we need. We then fit the model.
> library(dplyr)
> fpe <- read.table("https://grodri.github.io/datasets/effort.dat") |>
+ mutate(effort_g = cut(effort, breaks=c(min(effort), 5, 15, max(effort)),
+ right=FALSE, include.lowest=TRUE, labels=c("Weak","Moderate","Strong")))
> m2cg <- lm(change ~ setting + effort_g, data=fpe) All of the diagnostic measures discussed in the lecture notes can be calculated in Stata and R, some in more than one way.
Residuals
Let us start with the residuals. The extractor function
residuals() returns raw residuals, and has an optional
argument to return other types, but I find it easier to use
rstandard() for standardized and rstudent()
for studentized residuals. Let us obtain all three:
> ddf <- data.frame( residuals=residuals(m2cg), rstandard=rstandard(m2cg),
+ rstudent=rstudent(m2cg))Leverage and Influence
To get the diagonal elements of the hat matrix and Cook’s distance we
use the extractor functions hatvalues() and
cooks.distance():
> library(dplyr)
> ddf <- mutate(ddf, hat=hatvalues(m2cg), cooks=cooks.distance(m2cg))We are now ready to print Table 2.29 in the notes.
> ddf residuals rstandard rstudent hat cooks
Bolivia -0.83227667 -0.168973754 -0.163754309 0.2616128 2.529026e-03
Brazil 3.42822888 0.657314248 0.645213030 0.1720945 2.245290e-02
Chile 0.44160541 0.083498910 0.080865093 0.1486769 3.044043e-04
Colombia -1.52718268 -0.291358059 -0.282857589 0.1637904 4.156879e-03
CostaRica 1.28794371 0.242731989 0.235458166 0.1431063 2.459947e-03
Cuba 11.44160541 2.163382852 2.490348474 0.1486769 2.043412e-01
DominicanRep 11.29992007 2.161597338 2.487444785 0.1682585 2.363079e-01
Ecuador -10.03861524 -1.925295856 -2.126718448 0.1725535 1.932498e-01
ElSalvador 4.65406134 0.895661609 0.889814268 0.1782050 4.348948e-02
Guatemala -3.49960036 -0.685374906 -0.673572655 0.2064620 3.055405e-02
Haiti 0.02966757 0.006930347 0.006710289 0.4422478 9.520821e-06
Honduras 0.17747027 0.035544869 0.034417531 0.2412746 1.004433e-04
Jamaica -7.21985927 -1.361728588 -1.402245438 0.1444142 7.824693e-02
Mexico 0.90481996 0.183036697 0.177410357 0.2562359 2.885500e-03
Nicaragua 1.44383484 0.272655282 0.264612794 0.1465179 3.190539e-03
Panama -5.71205629 -1.076521261 -1.082268869 0.1431063 4.838569e-02
Paraguay -0.57177112 -0.109628999 -0.106187711 0.1720945 6.245643e-04
Peru -4.40250346 -0.841096436 -0.833012218 0.1661363 3.523718e-02
TrinidadTobago 1.28794371 0.242731989 0.235458166 0.1431063 2.459947e-03
Venezuela -2.59323608 -0.575229384 -0.562813507 0.3814295 5.100904e-02Here is an easy way to find the cases highlighted in Table 2.29, those with standardized or jackknifed residuals greater than 2 in magnitude:
> filter(ddf, abs(rstandard) > 2 | abs(rstudent) > 2) residuals rstandard rstudent hat cooks
Cuba 11.44161 2.163383 2.490348 0.1486769 0.2043412
DominicanRep 11.29992 2.161597 2.487445 0.1682585 0.2363079
Ecuador -10.03862 -1.925296 -2.126718 0.1725535 0.1932498We will calculate the maximum acceptable leverage, which is 2p/n in general, and then list the cases exceeding that value (if any).
> hatmax = 2*4/20
> filter(ddf, hat > hatmax) residuals rstandard rstudent hat cooks
Haiti 0.02966757 0.006930347 0.006710289 0.4422478 9.520821e-06We find that Haiti has a lot of leverage, but very little actual
influence. Let us list the six most influential countries. I will do
this using arrange() with desc() to sort in
descending order, and then using subscripts to pick the first 6
rows.
> arrange(ddf, desc(cooks))[1:6,] residuals rstandard rstudent hat cooks
DominicanRep 11.299920 2.1615973 2.4874448 0.1682585 0.23630787
Cuba 11.441605 2.1633829 2.4903485 0.1486769 0.20434122
Ecuador -10.038615 -1.9252959 -2.1267184 0.1725535 0.19324976
Jamaica -7.219859 -1.3617286 -1.4022454 0.1444142 0.07824693
Venezuela -2.593236 -0.5752294 -0.5628135 0.3814295 0.05100904
Panama -5.712056 -1.0765213 -1.0822689 0.1431063 0.04838569Turns out that the D.R., Cuba, and Ecuador are fairly influential observations. Try refitting the model without the D.R. to verify what I say on page 57 of the lecture notes.
Residual Plots
On to plots! Here is the standard residual plot in Figure 2.6, produced using the following code:
> library(ggplot2)
> ddf <- mutate(ddf, fitted = fitted(m2cg))
> png("fig26r.png", width=500, height=400)
> ggplot(ddf, aes(fitted, rstudent)) + geom_point() +
+ ggtitle("Figure 2.6: Residual Plot for Ancova Model")
> dev.off()png
2 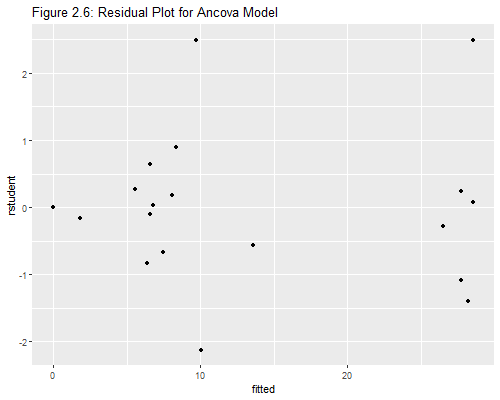
Exercise: Can you label the points corresponding to Cuba, the D.R. and Ecuador?
Q-Q Plots
Now for that lovely Q-Q-plot in Figure 2.7 of the notes:
> library(ggplot2)
> png("fig27r.png", width=500, height=400)
> ggplot(ddf, aes(sample = rstudent)) +
+ stat_qq() + geom_abline(intercept=0, slope=1) +
+ ggtitle("Figure 2.7: Q-Q Plot for Residuals of Ancova Model")
> dev.off()png
2 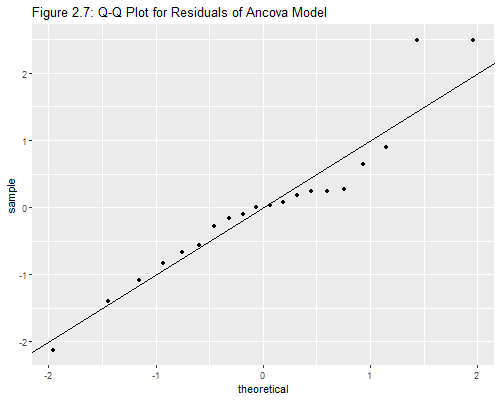
Wasn’t that easy? The qnorm() function in base R will do
this plot, but I used the equivalent stat_qq() in the
ggplot2 package. I superimposed a 45 degree line rather
than using stata_qq_line(). It is not clear to me how they
approximate the rankits, but the calculation seems very close to
(i-3/8)/(n+1/4), except for a couple of points on either tail.
Of course you can use any approximation you want, albeit at the expense
of additional work.
Filliben
I will illustrate the general idea by calculating Filliben’s
approximation to the expected order statistics or rankits. After sorting
the studentized residuals I use row_number() for the
observation number and nrow() for the number of cases.
> N <- nrow(ddf)
> ddf <- arrange(ddf, rstudent) |>
+ mutate(p = (row_number()-0.3175)/(N + 0.365))
> ddf$p[1] <- 1-0.5^(1/N); ddf$p[N] <- 0.5^(1/N)
> ddf <- mutate(ddf, filliben = qnorm(p))
> summarize(ddf, r_rstudent=cor(rstudent,filliben),
+ r_rstandard=cor(rstandard,filliben)) r_rstudent r_rstandard
1 0.9517735 0.9655174The correlation is 0.9518 using jack-knifed residuals, and 0.9655 using standardized residuals. The latter is the value quoted in the notes. Both are above (if barely) the minimum correlation of 0.95 needed to accept normality. I will skip the graph because it looks almost identical to the one produced above.
Updated fall 2022
